MBI Videos
Workshop 2: Control of Cellular and Molecular Systems
-
 Jeff Moehlis
Jeff MoehlisNonlinear oscillators - dynamical systems with stable periodic solutions - arise in many systems of physical, technological, and biological interest. Examples from biology include pacemaker cells in the heart, the firing of action potentials in neurons, and circadian rhythms. There are situations in which it is desirable to control biological oscillators, for example changing the phase of the circadian rhythm in order to adjust to a new time zone. With this in mind, we have developed an optimal control algorithm to change the phase of a periodic orbit using a minimum energy input, which also minimizes the transversal distance to the uncontrolled periodic trajectory. Our algorithm uses a two-dimensional augmented phase reduction technique based on both isochrons and isostables. This control algorithm is effective even when a large change in time period is required or when the nontrivial Floquet multiplier of the periodic orbit is close to one; in such cases, an analogous control algorithm based on standard phase reduction fails. Inspired by deep brain stimulation treatment of Parkinson's disease, we have also developed control algorithms for desynchronizing populations of oscillators, for example by maximizing the Lyapunov exponent associated with their phase dynamics, and through optimal phase resetting.
-
 Mary Ann Horn
Mary Ann HornDiacylgylcerol (DAG) plays a key role in cellular signaling as a second messenger. In particular, it regulates a variety of cellular processes and the breakdown of the signaling pathway that involves DAG contributes to the development of a variety of diseases, including cancer. A mathematical model of the G-protein signaling pathway in RAW 264.7 macrophages downstream of P2Y6 activation by the ubiquitous signaling nucleotide uridine 5’-diphosphate is presented. The primary goal is to better understand the role of diacylglycerol in the signaling pathway and the underlying biological dynamics that cannot always be easily measured experimentally. The model is based on time-course measurements of P2Y6 surface receptors, inositol trisphosphate, cytosolic calcium, and with a particular focus on differential dynamics of multiple species of diacylglycerol. When using the canonical representation, the model predicted that key interactions were missing from the current pathway structure. Indeed, the model suggested that to accurately depict experimental observations, an additional branch to the signaling pathway was needed, whereby an intracellular pool of diacylglycerol is immediately phosphorylated upon stimulation of an extracellular receptor for uridine 5’-diphosphate and subsequently used to aid replenishment of phosphatidylinositol. As a result of sensitivity analysis of the model parameters, key predictions can be made regarding which of these parameters are the most sensitive to perturbations and are therefore most responsible for output uncertainty. (Joint work with Hannah Callender, University of Portland, and the H. Alex Brown Lab, Vanderbilt.)
-
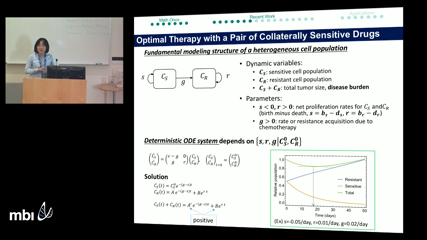 Nara Yoon
Nara YoonCancer is a disease developed by uncontrolled growth of mutated cells. To study such cancer, many different scales of researches has been carried out, from a small size of molecules to large size of organisms [1,2]. In this talk, I will give a brief overview about the range of mathematical oncology, and then talk about a recent project of a cellular scale modeling worked by my team.
In the project, we developed a model of ordinary differential equations and study the effect of sequential therapy on heterogeneous tumors comprised of resistant and sensitivity cells. Based on the model, we figured out (i) the optimal drug-switch strategy, and (ii) how composition of sensitive and resistant cell populations changes. Beyond our analytic results, we explored an individual based stochastic model and presented the distribution of extinction times for the classes of solutions found. Taken together, our results suggest opportunities to improve therapy scheduling in clinical oncology.
Reference
1. Anderson, A. R., & Quaranta, V. (2008). Integrative mathematical oncology. Nature reviews. Cancer, 8(3), 227.
2. Byrne, H. M. (2010). Dissecting cancer through mathematics: from the cell to the animal model. Nature reviews. Cancer, 10(3), 221.
-
 Ron Weiss
Ron WeissSynthetic biology is revolutionizing how we conceptualize and approach the engineering of biological systems. Recent advances in the field are allowing us to expand beyond the construction and analysis of small gene networks towards the implementation of complex multicellular systems with a variety of applications. In this talk I will describe our integrated computational / experimental approach to engineering complex behavior in mammalian cells. In our research, we appropriate design principles from electrical engineering and other established fields. These principles include abstraction, standardization, modularity, and computer aided design. But we also spend considerable effort towards understanding what makes synthetic biology different from all other existing engineering disciplines and discovering new design and construction rules that are effective for this unique discipline. We will discuss experimental results with synthetic biology building blocks for intracellular sensing, processing, and actuation in mammalian cells. We will then present a genetic circuit that can detect and destroy specific cancer cells based on the presence or absence or specific biomarkers in the cell. We will also discuss preliminary experimental results for obtaining precise spatiotemporal control over stem cell differentiation for tissue engineering applications. Finally, we will discuss a new framework for creating regulatory circuits based strictly on protein-protein interactions. These protein-phosphorylation based circuits operate at much faster speeds than existing transcriptional and translational based systems, with the ability to respond to a stimulus within a few seconds, thereby creating opportunities for new synthetic biology capabilities and applications.
-
 Gregory Batt
Gregory BattFeedback control methods have recently been applied to successfully take control of cellular functions. This novel field of research aims to remotely pilot cellular processes in real-time with unprecedented levels of robustness and precision to leverage the biotechnological potential of synthetic biology. Yet, the control of only a small number of genetic circuits has been tested so far. In this presentation, I will present the control of a multistable gene regulatory network, which is ubiquitously found in nature and play critical roles in cell differentiation and decision-making. Using an in silico feedback control loop, we demonstrate that a bistable genetic toggle switch can be dynamically maintained near its unstable equilibrium position. Thus, single cells could be controlled to remain in an undecided state for extended periods of time. Importantly, we show that a direct method based on dual periodic forcing is sufficient to simultaneously maintain many cells in an undecided state. These findings pave the way for the control of more complex cell decision-making systems at both the single cell and the population levels, with vast fundamental and biotechnological applications.
-
 Joerg Stelling
Joerg StellingSynthetic gene circuits have to operate in natural systems such as cells or organisms, with corresponding load on and cross-talk with them. These aspects are of particular relevance for biomedical applications where multiple design objectives with trade-offs (e.g., efficiency and robustness) and multiple scales (e.g., organism-wide impact of cell-based therapeutics) need to be considered. The first part of the talk will describe a Bayesian circuit design method that identifies circuit topologies whose behavior is robust to variations in parameters; it enables to reliably assess trade-offs between performance, robustness, and experimental feasibility, thus increasing the probability of success of circuit implementation. The second part will discuss a biomedical application of synthetic gene circuit design for in vivo closed-loop control, specifically for the treatment of type 1 and 2 diabetes; we achieved glucose responsiveness by a synthetic circuit that couples glycolysis-mediated calcium entry to an excitation-transcription system controlling therapeutic transgene expression. The examples help to argue that novel systems analysis methods are needed to enable efficient computational design of synthetic circuits, and how the design of synthetic systems allows us to refine our understanding of natural biological systems.
-
 Guy-Bart Stan
Guy-Bart StanIn this talk I will give an overview of some of our research activities in the "Control Engineering Synthetic Biology" group, where we focus our efforts on developing foundational forward-engineering methods to mathematically model, control, and experimentally implement synthetic gene circuits and cellular systems that aim at increasing the robustness, performance, and genetic stability of engineered cells. During the talk, I will propose some approaches to answer some of the following questions in systems and synthetic biology:
- How can we use cellular resources more efficiently to simultaneously improve growth rates and production yields?
- What is the interplay between feedback and buffering in cellular homeostasis?
- How can we create genetic oscillators for which the amplitude and period of oscillations can be tuned independently?
-
 Brian Ingalls
Brian IngallsAntibiotic-resistant pathogens present an increasing global health concern. Our group is investigating synthetic biology-based strategies for suppression of resistance in environmental bacterial populations. This approach involves the delivery of engineered genetic elements to target populations. We are developing models of the dynamics of this system, at both the genetic and population level, to be used for model-based design of potential implementations. Analysis of proof-of-principle scenarios and accompanying experimental results will be presented.
-
 Murat Arcak
Murat ArcakBreaking symmetry in spatially distributed systems is a fascinating dynamical systems problem and is of fundamental interest to developmental biology. In this talk we discuss several feedback mechanisms that enable formation of gene expression patterns in multi-cellular organisms. With the help of dynamical models we reveal the key structural properties that are necessary for patterning and present novel synthetic gene networks built upon these models.
-
 Johan Paulsson
Johan Paulsson
